Tsaousis lab (Laboratory of Molecular and Evolutionary Parasitology) current research is focused on the investigations of the adaptations of microbial eukaryotic organisms, and their course in parasitic evolution and diversity. To accomplish this, our laboratory is combining detailed bioinformatics analyses of newly generated genomic/transcriptomic/metabolomic results with field, cell biological and biochemical methods to investigate the parasitic and free-living microbial eukaryotes living in diverse and extreme environments.
Current projects:
Cryptosporidium and cryptosporidiosis
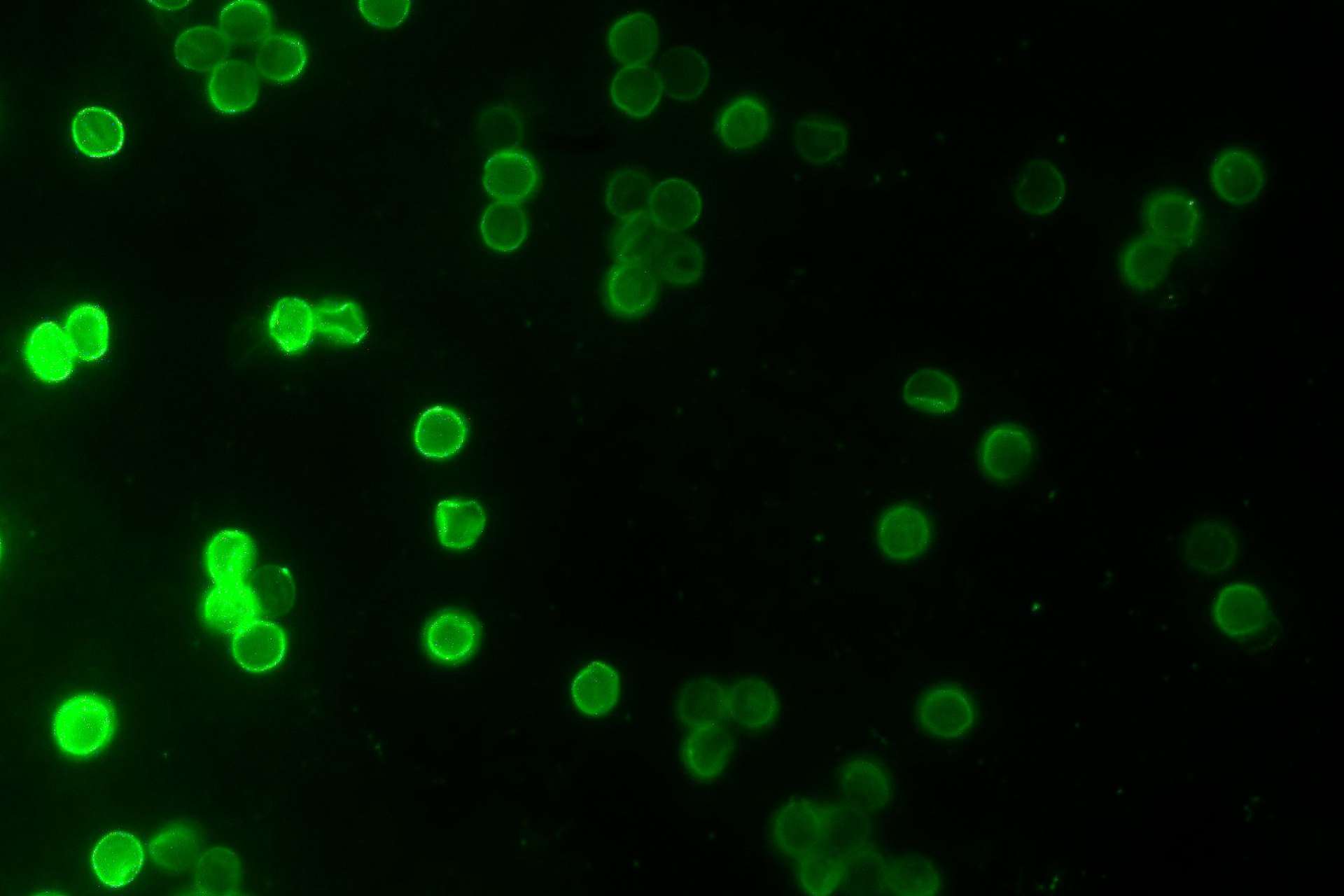
Cryptosporidiosis is a diarrhoeal disease caused by Cryptosporidium, a pathogen of great medical importance, which has appeared in the headlines several times in the past decades. An important fact about cryptosporidiosis is the lack of medical treatment in the form of drugs or vaccines. The parasite is mainly affecting children of a young age (below five), but people with impaired immune systems are also at great risk. In some cases, infected individuals have to deal with unpleasant diarrhoea lasting for several weeks, leading to dehydration that could potentially be deadly. Information on the infection patterns of the parasite and its interactions with the host is very limited, due to the lack of a laboratory system that will enable us to monitor the infection and replication processes of Cryptosporidium within a cell. Our laboratory has managed to overcome this difficulty by infecting with Cryptosporidium different types of cancer cells in a laboratory setting and testing whether they could successfully allow the parasite to grow and replicate. Our laboratory is currently interested to investigate which metabolites the parasite steals from the host cell and how it manipulates the molecular mechanisms of the host for its benefit. In addition, we are currently examining the cellular geography of Cryptosporidium inside and outside its host and the evolutionary adaptations that make it such a successful parasite. Our work will demonstrate what the molecular interactions between Cryptosporidium and its host are, will provide a better understanding of how complex the life-cycle of the parasite is and will generate essential knowledge about this medically important pathogen and will provide new targets for anti-parasitic drug development.
Establishing Naegleria as a model system to investigate adaptations to eukaryotic cellular adaptations
This project aims to develop tools and use them to study an organism that is neither animal, plant, algae, nor parasite. It is a single-celled creature living in soils and freshwater around the world. This creature, Naegleria gruberi, possesses nearly all of the cellular features found in animal and plant cells, but evolved away from them nearly 1.5 billion years ago. It is a uniquely placed sampling point from which to collect information about how cells work and gain a global perspective applicable to all eukaryotic cells. Our laboratory is currently developing a state-of-the-art genome editing system based on CRISPR/Cas9 based methodology. This will enable the systematic interrogation of Naegleria’s genome function by making this organism amenable to the full array of CRISPR-based approaches currently established for other organisms, including knockout, knockdown and activation screens of various genes. We will aim to develop this approach into a powerful high-throughput functional genomics toolbox, thereby enabling understanding of the function(s) of the 15,727 protein-coding genes that are present in Naegleria’s nuclear genome. The overall outcome of this project is to produce a set of protocols, plasmids and tools to be used by the scientific community to address diverse scientific questions, using Naegleria gruberi as a model system.
Exploring the anaerobic and other unique adaptations of Blastocystis
Blastocystis is an obligate anaerobic parasite also found in patients with irritable bowel syndrome. The actual pathogenicity of Blastocystis is still questionable, since currently there is no direct link between the parasite and the disease caused. As an anaerobic organism, Blastocystis harbor peculiar Mitochondrion-related organelles (MROs), which are considered to be an intermediate form between a typical mitochondrion and a hydrogenosome. Another interesting fact about Blastocystis, concerns the presence of peculiar proteins encoded from its genome: it seems that Blastocystis is a “lateral gene transfer magnet” since several genes have been acquired from diverse eukaryotes and prokaryotes in order to assemble a kind of mixed genome. Using a combination of bioinformatics along with cellular and biochemical techniques, our laboratory aims to investigate these “novel” functions in Blastocystis and its closely relatives (e.g. Proteromonas) and attempt to understand their evolutionary history and the reason for their existence.
Host-parasite interactions in the gut and how parasites shape the host’s microbiome
Studies exploring the eukaryotic component of the gut microbiota lag far behind those of prokaryotes. Efforts in documenting eukaryotes from the human and animal gut are still at their infancy. Despite this, several reports have demonstrated a clear association between the eukaryotic residents of the gut microbiome with both genetic and neglected diseases in humans and animals. Our laboratory is currently investigating such associations using both genetic and metabolomics studies. For this work, we are currently collaborating with both the Wildwood Trust and Howletts Wildlife Parks to investigate and compare the gut microbiome and metabolome of free-living animals and those kept in captivity. Our laboratory is also part of the ThaiGut consortium that aims to investigate the role of the gut microbiota in health and disease in humans in Thailand and its neighbour countries.